-
Whitepaper
TAP® Empowers High-Quality Genome Studies with Illumina Infinium CytoSNP-850K Array
HIGHLIGHTS
- The TAP is a non-invasive and virtually painless blood collection device that enables point-of-care, high-quality whole blood collections for downstream molecular applications such SNP profiling by microarrays.
- The Illumina Infinium CytoSNP-850K array kit enables cost-effective and comprehensive genome-wide association studies (GWAS) for nearly 850,000 empirically selected variant markers spanning the genome, providing disease-focused coverage 3,262 genes of known cytogenetics relevance in both constitutional and cancer applications.
- (Data concordance assessments for microarray quality metrics (such as call rate/quality, gender sample QC, replication error rate, comparisons of theta, R-squared values, B allele frequencies, genotype bins, cluster analysis for automatic sample pairing detection, dendrogram analysis for likeness, human ID RS-site analysis for sample identification and genotype plotting of AA/AB/BB genotypes within the array) demonstrate variant call quality equivalence between TAP vs traditional venipuncture collections.
- Applications for high-throughput blood collection coupled to high-throughput SNP profiling enables efficient global GWAS for research, screening and diagnostic applications.

Figure 1: Overall workflow showing TAP blood collection through microarray analysis on GenomeStudio software.
INTRODUCTION
SNP profiling via the Illumina Infinium CytoSNP-850K array leverage international consortium data to identify qualified SNPs for a comprehensive view of the human genome. The coverage of the array is intentionally designed for cytogenetics, cancer, constitutional and other disease-related applications. The array provides high specificity of SNP targets by using 50-mer to increase sensitivity and resolution for regions of interest. This is especially important for copy number variations and mosaic detection. Redundancy is built into the chip, as each chip SNP provides at least 15x coverage per site. Altogether, the CytoSNP-850K array enables LOH, AOH, UPD, CNV and SNP-specific calls with high signal to noise ratio per site. In addition to SNP data capture, chromosomal and structural aberrations can be detected as a low-cost method for comprehensive genomic profiling.
Traditional blood collection for the Illumina microarray system involves a venipuncture draw by a trained phlebotomist, whereby 5-10 ml of whole blood is collected in a standard citrate/heparin/EDTA tube. This procedure is relatively costly and inaccessible to scale across large patient populations for point of care testing. Economic and logistical hurdles to high-quality blood collections persist due to the modality of the sample collection process (primarily due to the need of a trained phlebotomist/technician), thereby impeding access to many people for efficient point-of-care testing.
Here, we introduce the Yourbio Health TAP blood collection device as a viable tool to enable feasibility of blood collection scale and adoptability for downstream molecular testing on the Illumina Infinium CytoSNP-850K array. The TAP enables point-of-care collections by novice users, direct use by physicians/technologists in general practices, or collections in underserved/remote geographies. We obtained paired blood samples (TAP + traditional venipuncture) to analyze downstream Illumina data to demonstrate analytical concordance verification of SNP calls and other quality metrics.
MATERIALS AND METHODS
Under an IRB-approved protocol, four donors had paired blood samples collected by a trained phlebotomist (traditional venipuncture tube) and assisted-collections (TAP device) into heparin-coated tubes. The venipuncture tubes collected the full max fill volume of 3 ml by the phlebotomist, whereas the TAP devices collected between 300-600 ul of whole blood. All blood samples were collected and treated identically (same-day shipment to lab on ice packs during transit) in preparation for downstream DNA extraction and library preparation for WES. Briefly, DNA extraction was performed with the Qiagen QIAamp DNA blood mini kit (pn 51104) using 200 ul as whole blood input and 200 ul of elution volume. DNA QC with the Qubit Flex fluorometer using the Qubit DS BR assay kit. Any sample below 40 ng/ul was concentrated to 100 ul of original elution via Savant DNA 120 Speed Vac concentrator. Gel confirmation of product was conducted via E-gel agarose gel electrophoresis system. The entire microarray process spanned 4 days and followed the Illumina microarray methylation protocol (see references for SOP). Results for 847, 140 rsIDs for SNP calls throughout the entire human genome were analyzed in a pairwise fashion between TAP vs VP for each donor using GenomeStudio 2.0 software (genotyping module).
RESULTS
The table shows DNA extraction Qubit results. Note that sample 31 and 32 were originally mislabeled on the collection tube, whereby identification of the error was found via SNP results and confirmation is demonstrated elsewhere (see STR Analysis white paper). The correct labeling of samples is shown in the table. All samples passed QC and moved forward with the microarray protocol.
Without initially linking any paired donor samples, a reproducibility error analysis was conducted on all 8 samples by running the reproducibility error algorithm to identify the replication error rate across all 8 samples. The desired outcome is such that matching donor samples (i.e. TAP and VP) should have matching replication error rates for all analyzed SNPs. The reproducibility error rate is calculated as 1 – sqrt(1 – errors/max_possible_errors).
The call rate demonstrates the percentage of SNPs that have a GenCall score greater than the automatically set threshold, highlighting the SNP callability for each sample. Additionally, an automated gender caller predicted the correct gender for each sample based on X and Y chromosomal SNPs within the 850K array. Overall, these initial parameters demonstrate high-quality DNA extraction and QC and correctly identified matching pairs of donor samples (TAP + VP).
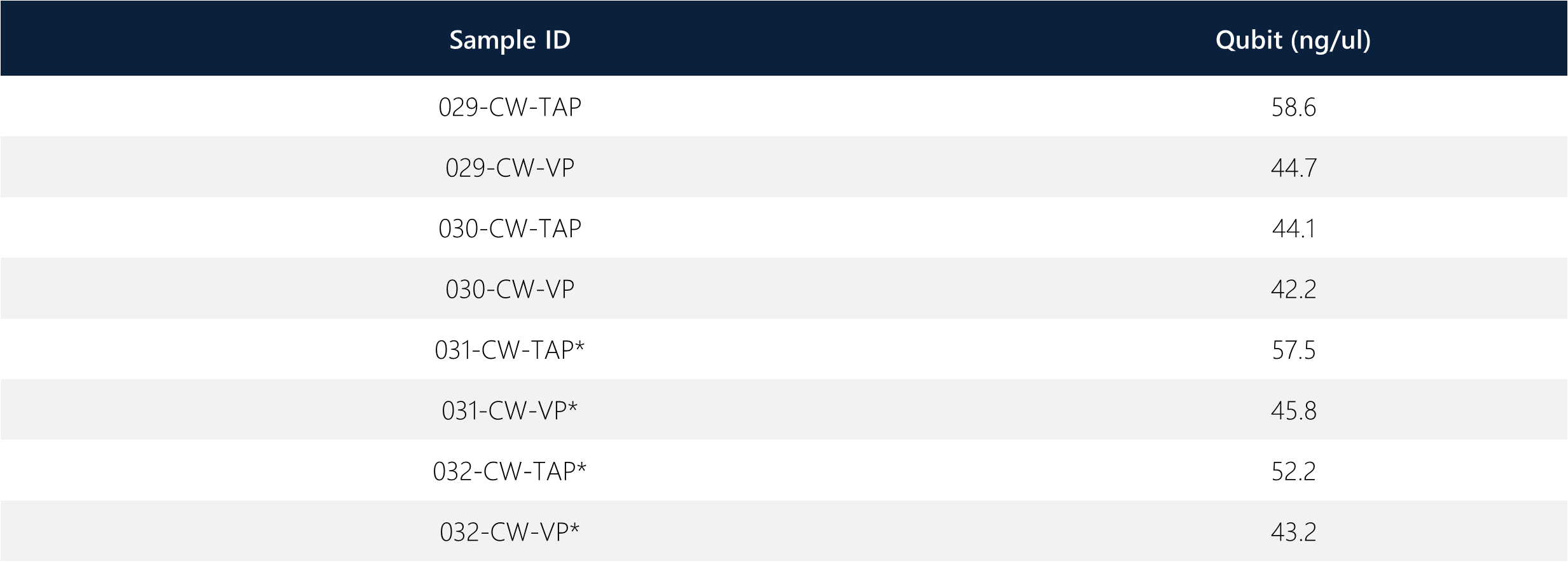
Table 1: DNA Qubit reading for all samples.
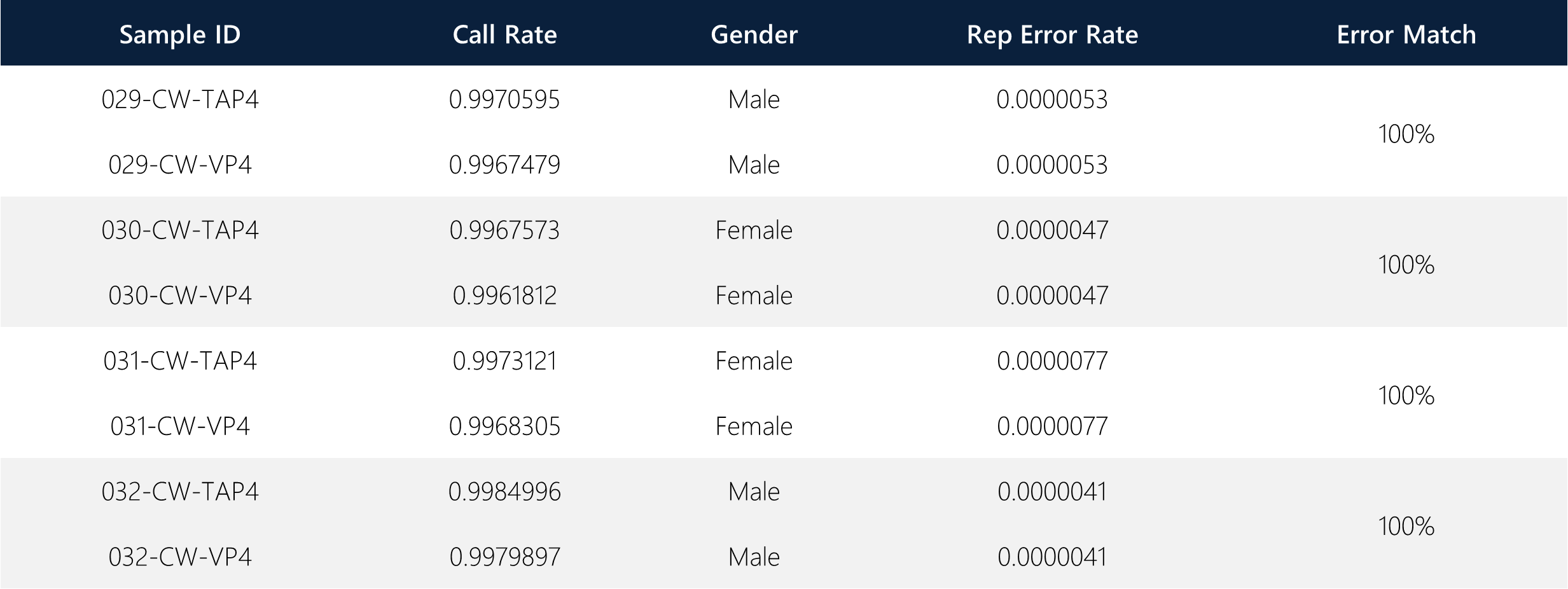
Table 2: Call rate, gender caller and replication error rates for all samples.
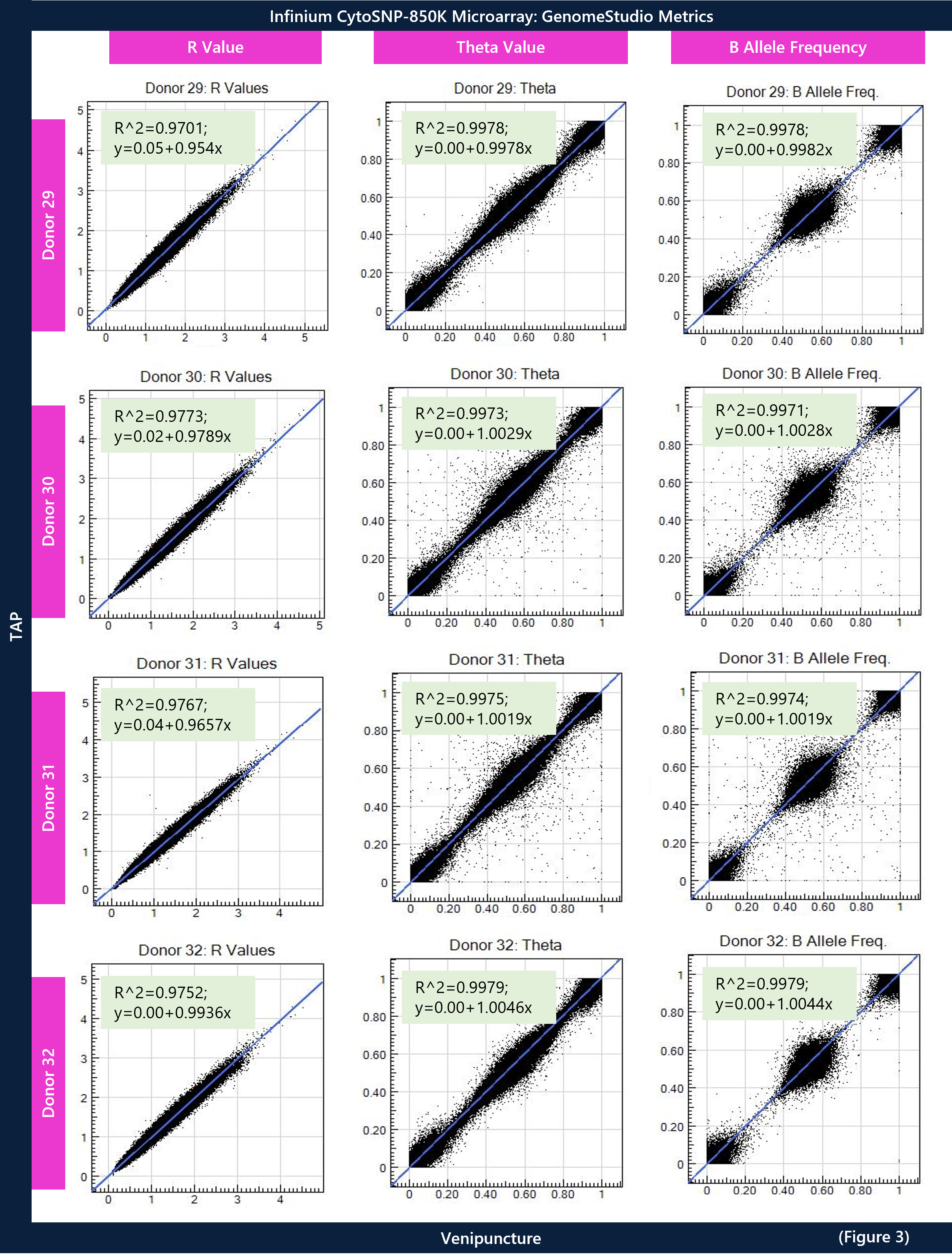
To demonstrate concordance between SNP calls, paired sample analysis for the VP sample (reference) was plotted against the TAP sample (subject). Theta values range from 0 to 1 and represent the fraction of bases that are genotyped as the B allele. For example, 0 translates to homozygous reference (AA), 0.5 for heterozygous (AB) and 1 for homozygous variant (BB). The R value represents the fluorescence intensity of the probe on the array for a sample. The B allele frequency for a sample is interpolated from known B allele frequencies of 3 canonical clusters: 0, 0.5, 1. Therefore, theta and B allele frequency values generated from GenomeStudio for each SNP call was plotted to show concordance between the VP and TAP samples from each donor. These values should theoretically have the same R^2 to demonstrate fidelity. The normalized R values were plotted for each donor to show correlation of fluorescence between TAP and VP. Lastly, the genotype bins for the VP sample were numerically assessed against the B allele frequency for the TAP sample to show any variance of calls between samples and the “no call” rate.
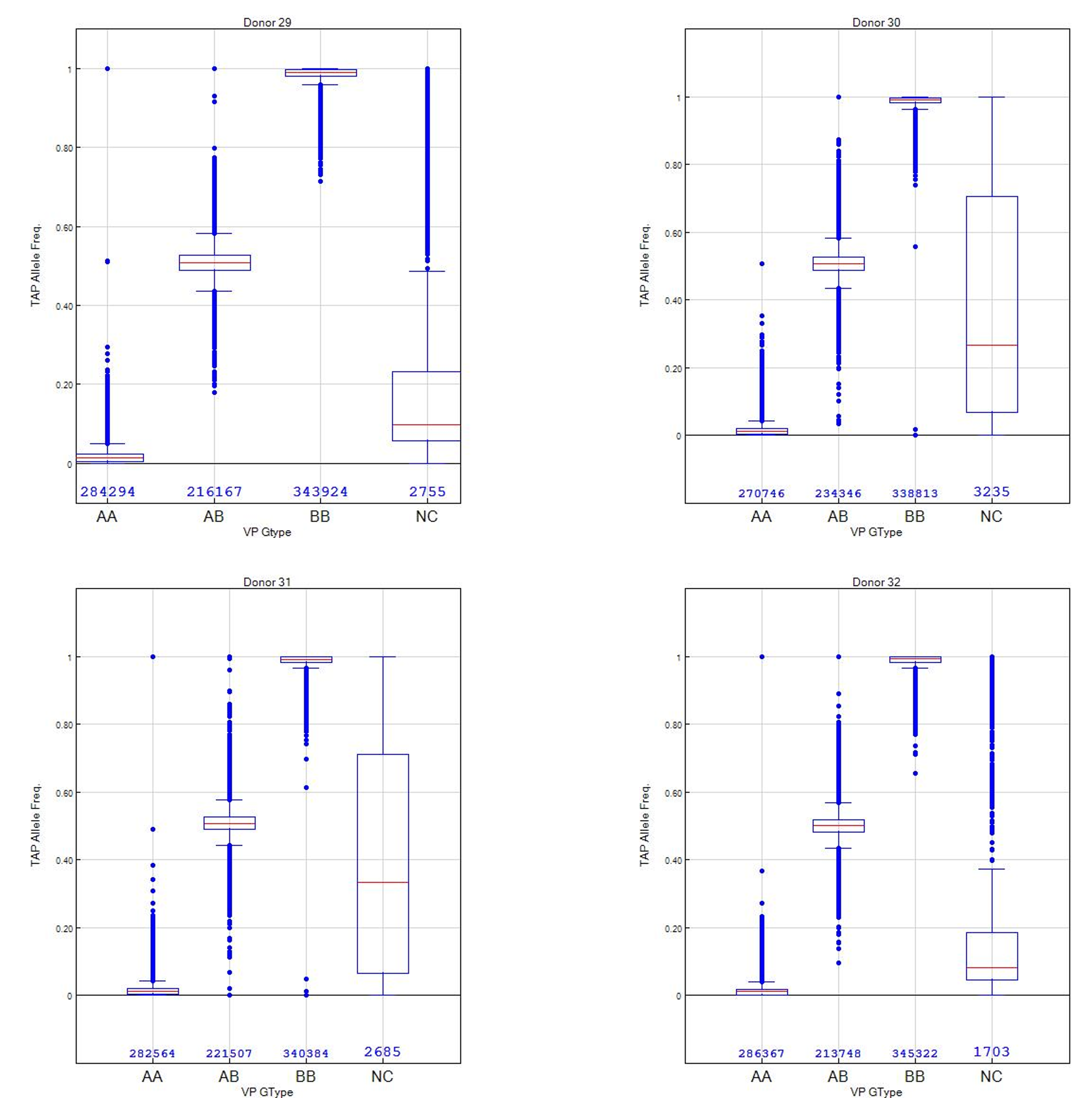
Figure 2: Box plot to categorize genotype bins for the VP sample (reference/truth) vs. the TAP B allele frequency to demonstrate constricted inter-quartile ranges for AA, AB and BB genotypes. Numerical counts (total 850K+) and the no-call rate is shown for context.
Without initial pairing, a Manhattan clustering algorithm was run within GenomeStudio to automatically identify paired donor samples based on SNP concordant calls. All TAP+VP samples from respective donors were matched with 100% identity across >850K+ SNPs. A snapshot of a heatmap and the clustered dendrogram is presented to show pairing of samples and genotype concordance (B allele frequency) for each donor pair. The complete heat map can be expanded to all ~850K SNPs, whereby clustering within genes, chromosomes and panels are possible.
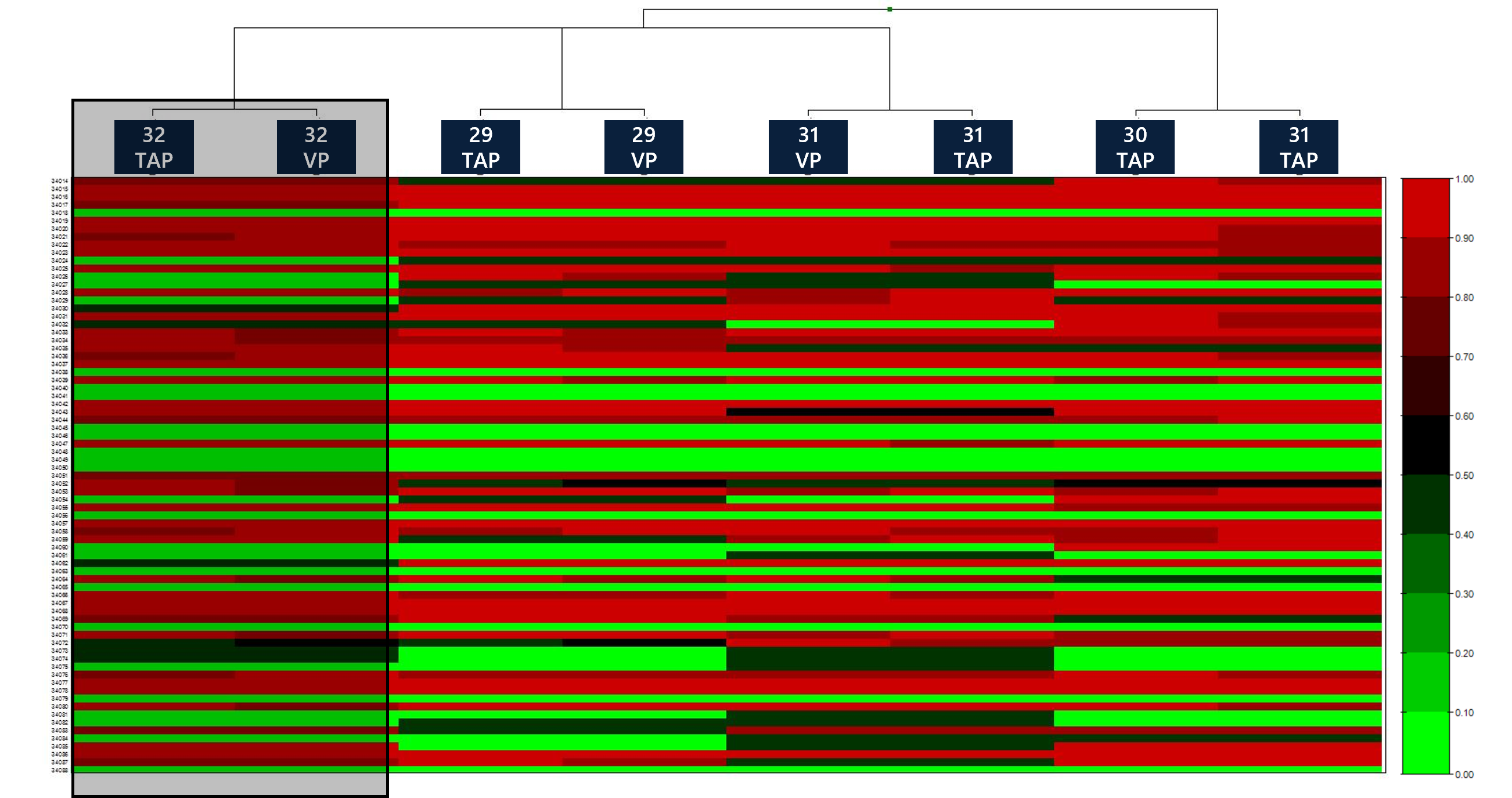
Figure 4: Example of heatmap showing all auto-clustered paired samples. One paired sample cluster (overarching yellow shade) showing matched color coordination of B allele frequencies (right y-axis) of SNP positions (left y-axis) between individual sample columns is highlighted for context.

Figure 5: Example of Y-chromosome ideogram mapped to SNP positions within the CytoSNP-850K microarray. Differences between male and female samples are shown.
CONCLUSIONS
Initial feasibility for the Illumina Infinium CytoSNP-850K microarray assay demonstrates good correlation of >850K SNP sites between TAP and VP samples. Specifically, the R^2 values for TAP and VP across all donors is >0.99, with 100% matching R^2 values for theta values vs. b allele frequencies between matrices. The genotype bins demonstrate very low IQR, with <0.3% no-call genotypes. Overall, the call rate across all samples is >99.6%, demonstrating good genotypic quality scores. Accessible and minimally invasive sampling with the TAP device can enable comprehensive genomic testing over more frequent timepoints for monitoring needs. Remote collections with TAP can enable access, throughput and disease monitoring for changes over time when coupled to global genomic-wide association assessment platforms such as the Illumina Infinium CytoSNP-850K microarray. The purpose of this study is to demonstrate technology and platform capability of the TAP device with the Illumina Infinium platform. Further evaluation of the TAP device could benefit from a larger sample size and comparisons to similar orthogonal assessments (for example, WES, WGS, low-pass sequencing and targeted sequencing). Future studies will explore the cytogenetic relevance of abnormal/disease vs. normal/reference populations. Additional considerations may include expansion of the microarray targets to more comprehensive SNP arrays (i.e. >2M SNP chips).