-
Whitepaper
Next Generation Carrier Screening on ThermoFisher Ion Torrent with TAP®
HIGHLIGHTS
- The TAP is a non-invasive and virtually painless blood collection device that enables point-of-care, high-quality whole blood collections for downstream NGS applications such as carrier screening.
- The Ion Torrent CarrierSeq Expanded Carrier Screening (ECS) enables an automated solution for expanded carrier screening (ECS) of 420 genes for inherited and germline disorders of autosomal recessive and X-linked conditions.
- Assessment of carrier sequencing quality metrics and variant calls via NGS demonstrate comparable equivalence between TAP vs traditional venipuncture blood collections, yielding 100% concordance of variant calls and 100% pass rate for acceptable QC criteria.
- Applications for rapid and high-throughput on-demand carrier screening via point of care blood collections enable global access to preconception, prenatal, postnatal and global population screening.
INTRODUCTION
Germline carrier screening is conducted for the assessment of specific genetic disorders, whereby a genetic variant associated to a pathogenic condition may propagate into future generations. While traditional targeted carrier screening is focused on a small number of prevalent diseases, a better approach is the use of an expanded carrier screening for a comprehensive assessment of multiple genes and variants from a single blood sample. Such diseases include Tay-Sachs, sickle-cell anemia, xeroderma pigmentosa, progeria and more. Autosomal recessive traits are particularly suited for this type of analysis because a large fraction of “carrier” individuals have no known prior family history of the condition. Next-generation sequencing (NGS) can enable comprehensive carrier screening for the identification of more at-risk individuals, including pregnant women, adults and children.
The traditional blood collection method for comprehensive, high-quality DNA sequencing involves a venipuncture by a trained phlebotomist, whereby 8-10 ml of whole blood is collected in a standard heparin/EDTA tube. This procedure is relatively costly and inaccessible to scale across large patient populations. Economic and logistical hurdles to high-quality blood collections persist due to the modality of the sample collection process, thereby impeding access to many people for precision medicine and ultimately decreasing the sample size for scientific and statistical power in clinical research.
Here, we introduce the Yourbio Health TAP blood collection device as a viable tool to enable comprehensive and expanded carrier screening. The TAP enables point of care collections by novice users or directly by physicians/technologists in general practices. We obtained paired blood samples (TAP + traditional venipuncture) to analyze downstream NGS data to demonstrate analytical concordance verification of sequencing variant calls and NGS quality metrics. We assess the quality of reads, coverage, throughput, read length, on-target rate, mapped reads, mean depth, coverage uniformity, DNA extraction QC and more.
MATERIALS AND METHODS
Under an IRB-approved protocol, six donors had paired blood samples collected by a trained phlebotomist (traditional venipuncture tube) and self-collected (TAP device) into heparin-coated tubes. The venipuncture tubes collected the full max fill volume of 3 ml by the phlebotomist, whereas the TAP devices collected a minimum of 300 ul of whole blood. All blood samples were collected and treated identically (room temperature storage and ambient transit) in preparation for downstream NGS. Briefly, library generation, template preparation, and sequencing were conducted via the Ion Torrent CarrierSeq Expanded Carrier Screening (ECS) kit, whereby prepared libraries were loaded on to Ion Chips and subsequently sequenced on the Ion GeneStudio S5 System.
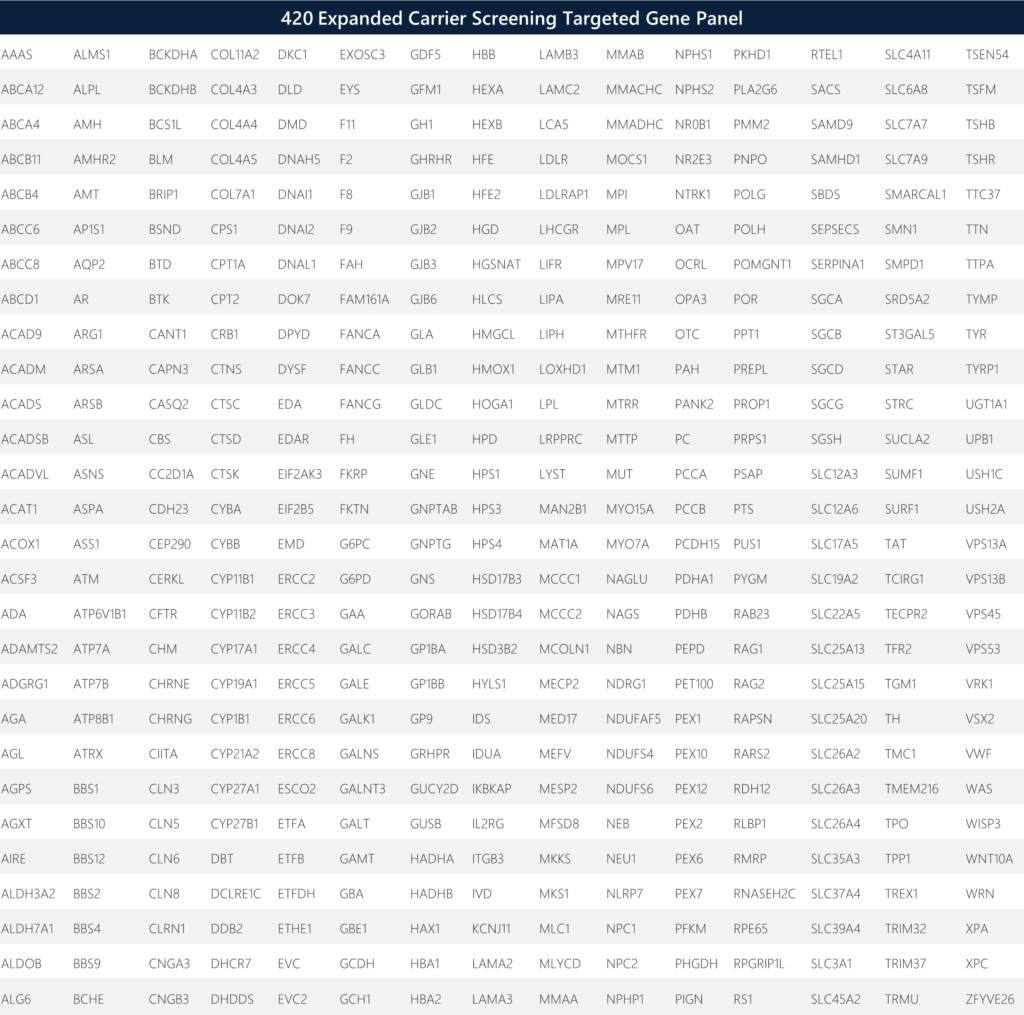
We chose the Thermofisher Expanded Carrier Sequencing panel via Ion AmpliSeq targeted sequencing as an end-to-end solution (sample to report) to analyze 14,044 amplicons covering the coding regions (CDS) of 420 genes, including +/-25 bp flanking intron/exon boundaries, as well as selected intergenic, intronic, and homologous regions. Note that the CDS regions are defined either by a specific transcript or a combination of multiple transcripts. The targeted regions are sequenced with the aim to achieve a uniformity of ≥93%, a q20 mean read length of >155 basepairs, and coverage of >200X when the reads are aligned to human genome assembly GRCh38 (hg38). Targeted regions assess the potential of >36,000 putative carrier single nucleotide archive of human variation and privately curated non-public variant sources. Variant calling is subject to quality control metrics, which include low read coverage. Variant calling of indels is limited in regions of homopolymer lengths of greater than eight nucleotides. Variant detection issues are possible in regions with low sequence complexity, large regional copy number changes, large indels, and regions with high homology to other genomic loci. Detection rates are ultimately determined based on analytical sensitivity, literature estimates for the disease allele contribution, and population frequency predictions. If variants have not been previously described in the literature, the detection rate might not be reported. Further, detection rates do not take into account the disease-specific rates of de novo mutations.
RESULTS
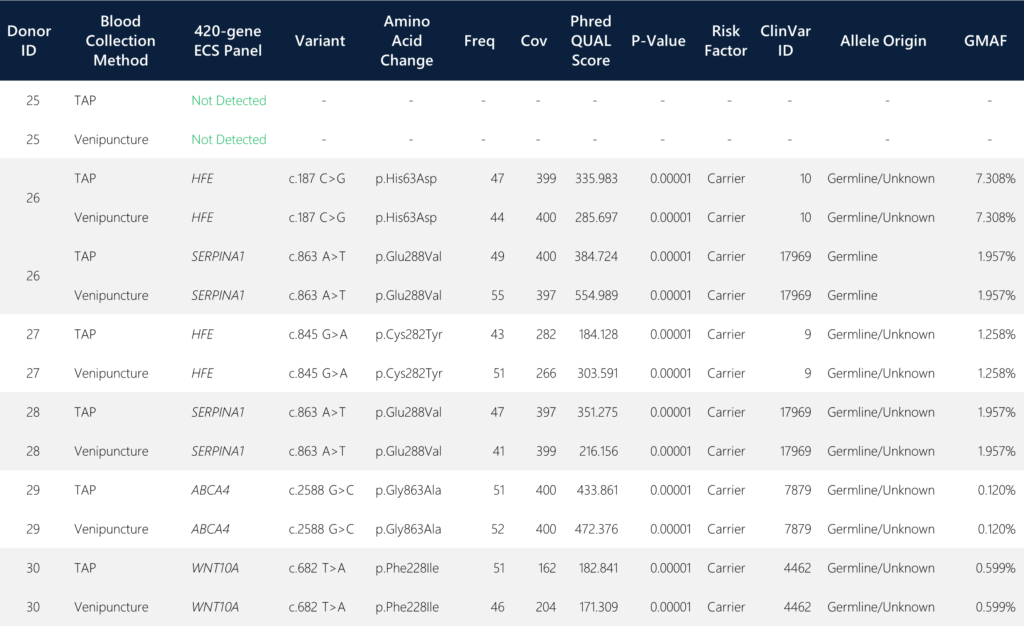
When screening for the targeted putative carrier variants covering 420 genes, we detected four genes with identified carrier alleles that were 100% concordant in variant calls between the TAP and the traditional venipuncture samples. The four detected genes include HFE (Homeostatic Iron Regulator), SERPINA1 (serine peptidase inhibitor alpha-1), ABCA4 (ATP-binding cassette subfamily A member 4), and WNT10A (Wnt Family Member 10A). All six paired sample sets yielded identical SNV call determinations (Table 1).
Additional NGS quality metrics were assessed for QC concordance and acceptance criteria. Overall, twelve donor samples, one blank, one positive control, one negative control and one no-template-control (NTC) were loaded onto a 16-sample Ion Chip for sequencing. Over 100 million total reads were obtained with 94% loading density of Ion Sphere Particles (ISP), representing 72% of usable reads after several high-quality filter assessments (exclusion of polyclonal, low quality and adapter dimer reads).
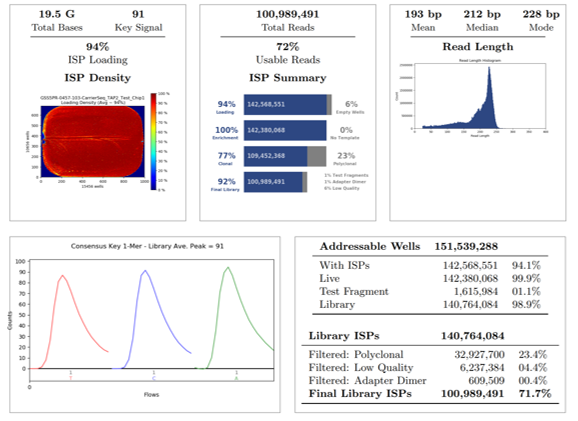
The final library was 92%, which represents the percentage of reads which pass all stringent quality filters and which are recorded in the BAM file. The final library ISPs is 72%, which represents the percentage of Library ISPs of reads passing all quality filters, also recorded in the BAM file. The read length histogram shows the trimmed lengths of all reads present, whereby the median read length in this run of combined samples is 212 bp.
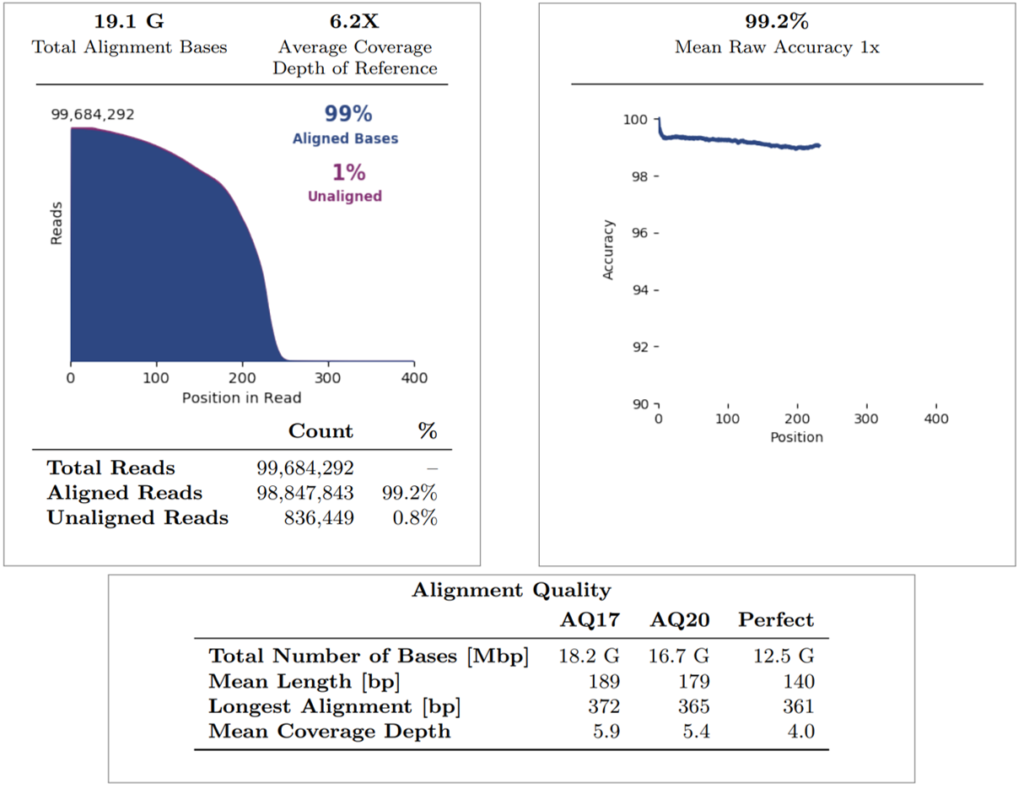
Regarding the alignment summary, the average coverage depth of reference was 6.2X, whereby all reads were aligned to GRCh38.p2 (note that internal Mask1 was applied and the reference coverage does not consider enrichment). The overall percentage of aligned bases and aligned reads is 99% for both. The mean raw accuracy at 1X is 99.2%, which represents the average raw accuracy of 1-mers plotted by their position in the reads. The alignment quality summary shows several quality metrics (AQ17, AQ20, Perfect), whereby the mean coverage depth is displayed at each quality level (note that this does not consider enrichment of the reference).
For sample level analysis and concordance assessments between both collection methods per donor, we compared metrics for mapped reads, on-target rate, mean depth of coverage (target enrichment), percentage uniformity, and the 20X coverage rate (not shown; all 100%). These assessments were specifically conducted on the targeted regions and sequences as described in the Materials and Methods section.
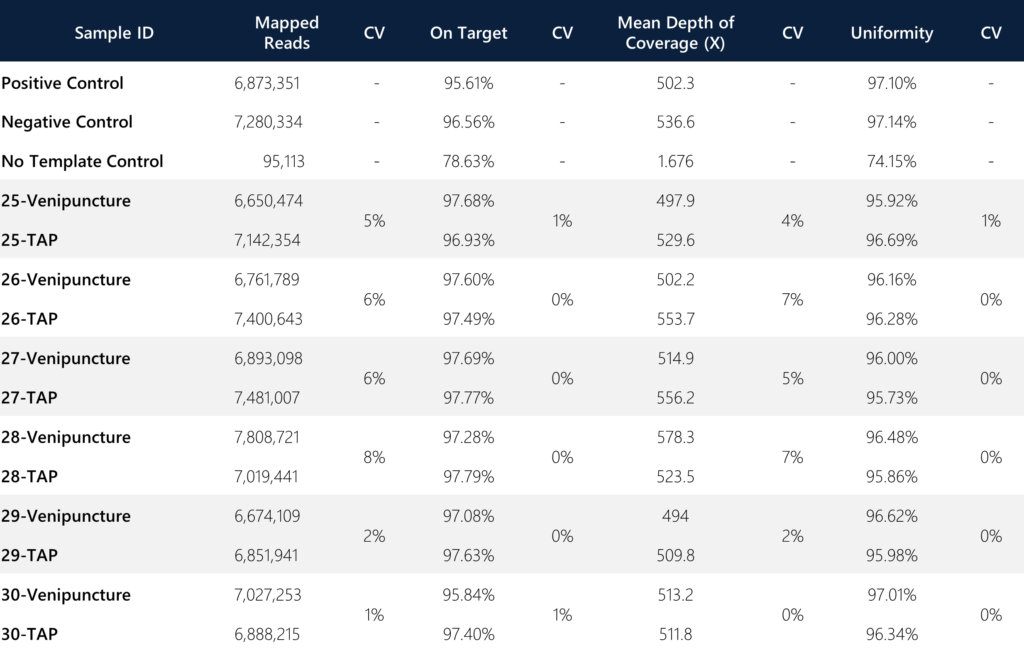
Of note, an important metric for consideration is the variance between the on-target rate and the uniformity of targeted regions between the sample types. In both instances, the variance was ≤1% between the TAP and venipuncture samples, indicating the degree of comparable quality for reads that are on-target (defined as the percentage of reads mapped to the targeted region relative to all reads mapped to the reference) and uniformity (defined as the percentage of bases in all targeted regions covered by at least 0.2X the average base read depth). The combined average of mean depth of coverage and the mapped reads for TAP was slightly better than venipuncture (Coverage: 530.77X vs. 516.75X, CV = 1.89%; Reads: 7,130,600 vs. 6,969,241, CV = 1.62%). Additionally, the overall average coverage across all paired samples (n=12) is 523.76X (for comparison, most target-enriched gene panels generally incorporate a minimum of 100X standard coverage as acceptance criteria).
CONCLUSIONS
Initial feasibility for the ThermoFisher ECS Ion Ampliseq platform shows concordance of variant outcomes and good quality metrics between TAP and VP collection methods. The purpose of comprehensive and expanded carrier screening is to be inclusive for pan-ethnic and diverse populations while significantly reducing risk via molecular interrogation of a large array of non-benign risk alleles for carrier status. Variants of pathogenic significance for inherited disorders and rare diseases may propagate into subsequent generations, even after conventional and traditional methods for screening which are limited in assessments. The first step towards mass population carrier screening for germline recessive diseases is the sample collection process, whereby inclusion of global populations is amenable from high-quality, point-of-care blood collection devices that overcome the access barrier and economic limitations of traditional phlebotomy. The TAP device enables efficient blood collection by lay-users, whereby the collected sample can be integrated into the streamlined end-to-end ThermoFisher ECS Ion Ampliseq platform for automated extraction, template and library preparation, variant analysis and annotation, and reporting with a single consolidated NGS screening workflow. This allows for both consumers and researchers to increase uptake of carrier screening with lower hurdles to access. We demonstrated 100% variant call concordance and ≤1% variance for on-target rate and uniformity in this pilot study while demonstrating various substantially equivalent QC metrics between sample matrices. Future studies will incorporate VCF concordance and comparisons across both sample types. Additional evaluations may include patient populations harboring variant classes of pathogenic, likely pathogenic, benign and variants of unknown significance, whereby detection and classification of performance by the ECS platform can be further assessed.
